The Application of Electron-electron Double Resonance (DEER) Technique in DNA Structure Analysis
Since the discovery of the classic double helix structure of DNA by Watson and Crick in the 1950s, DNA has become the core of life science research. The number and arrangement of the four bases in DNA lead to genetic diversity, and its spatial structure affects gene expression.
In addition to the traditional DNA double helix structure, a special four-stranded DNA structure called G-quadruplex has been discovered in human cells. G-quadruplex is a higher-order structure formed by the folding of DNA or RNA rich in tandem repeats of guanine (G). G-quadruplexes are highly abundant in rapidly dividing cells, such as cancer cells. Therefore, G-quadruplexes can serve as drug targets in cancer research. Investigating the structure of G-quadruplexes and their binding modes with ligands is of great significance for the diagnosis and treatment of cancer cells.
Electron-electron Double resonance (DEER)
Electron-electron double resonance (DEER) using pulsed dipolar electron paramagnetic resonance (PDEPR) has been developed as a reliable and versatile tool for structure determination in structural and chemical biology. DEER combined with site-directed spin labeling (SDSL) techniques can provide distance information at the nanoscale. In the study of G-quadruplex structures, DEER technology combined with SDSL can differentiate different lengths of G-quadruplex dimers and reveal the binding modes of G-quadruplex ligands with dimers.
PDEPR techniques can distinguish different lengths of G-quadruplex dimers.
The spin-label used for distance measurements in DEER experiments is Cu(pyridine)4. The Cu(pyridine)4 complex is covalently bound to G-quadruplexes, and the dipole-dipole interactions between two paramagnetic Cu2+ ions in the π-stacked G-quartet monomers can be measured. This allows for the study of dimer formation.
[Cu2+@A4] (TTLGGG) and [Cu2+@B4] (TLGGGG) are two oligonucleotides with different sequences. Figure 1 and Figure 2 show the DEER experimental results of [Cu2+@A4]2 and [Cu2+@B4]2, respectively. From the DEER results, the average distance between individual Cu2+-Cu2+ ions in [Cu2+@A4]2 dimer is dA = 2.55 nm. The G-quadruplexes at the 3′ ends of the G-quartets form G-quadruplex dimers through tail-to-tail stacking, and the gz axes of the two Cu2+ spin labels in the G-quadruplex dimer are arranged in parallel.
Compared to the [Cu2+@A4]2 dimers, the π-stacking distance in [Cu2+@B4]2 is longer (dB-dA = 0.66 nm), confirming the presence of an additional G-quartet in each [Cu2+@B4] monomer, which is consistent with the expected distance. Therefore, DEER measurements can differentiate different lengths of G-quadruplex dimers.
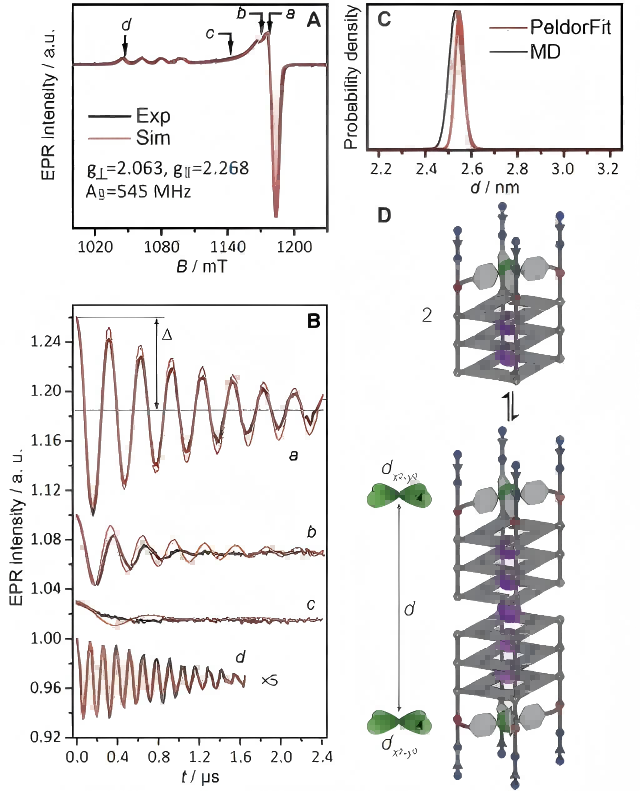
Figure 1 (A) Pulsed EPR spectrum (black line) of the [Cu2+@A4]2 dimer and its corresponding simulation (red line) (34 GHz, 19 K); (B) Background-corrected DEER time-domain traces (black line) at four field positions (a-d) and the best-fit results obtained from PeldorFit (red line); (C) Distance distribution obtained using PeldorFit (red line) and MD simulation (gray line); (D) Equilibrium between the monomer [Cu2+@A4] and the dimer [Cu2+@A4]2. (Angew.Chem. Int. Ed. 2021, 60,4939 –4947)
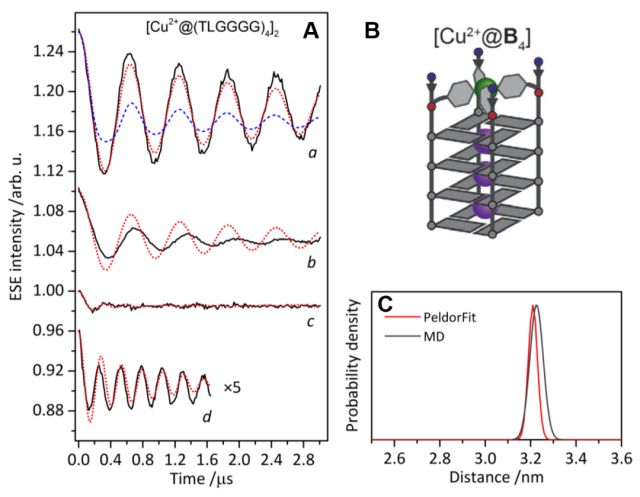
Figure 2 (A) shows the background-corrected DEER time domain plots (black lines) at four different field positions (a-d) for [Cu2+@B4], along with the best fit obtained from PeldorFit (red lines). (B) is a schematic representation of the [Cu2+@B4] structure. (C) represents the distance distribution obtained using both PeldorFit (red line) and MD simulation (gray line). (Angew. Chem. Int. Ed., 2021, 60, 4939-4947)
Exploring the Binding Mode of G-Quadruplex Ligands with Dimers Using DEER Technique
Many small molecules and metal complexes with planar aromatic conjugated systems and positive charges can bind and stabilize folded secondary structures, making them potential anticancer drugs.
N, N’-Bis[2-(1-pyridyl)ethyl]terephthalamide (PIPER) is a well-known G-quadruplex binder that can interact and stabilize G-quadruplexes through π-stacking interactions. DEER technology can be used to investigate the binding modes of PIPER with G-quadruplexes.
Figures 3 and 4 show the DEER experimental results of different ratios of PIPER to [Cu2+@A4]2 dimer. The results show that when the ratio of PIPER to [Cu2+@A4]2 dimers is 1:1 (PIPER@[Cu2+@A4]2), dP = 2.82 nm.
Compared to the pure [Cu2+@A4]2 dimers (dA = 2.55 nm), the distance between Cu2+-Cu2+ ions increases, indicating the formation of a sandwich complex where the planar organic molecules are inserted between the 3′ faces of the two G-quartet monomers. When the ratio of PIPER to [Cu2+@A4]2 dimer is 2:1 (2PIPER@[Cu2+@A4]2), d2P = 3.21 nm.
Compared to PIPER@[Cu2+@A4]2 dimer (dP = 2.82 nm), an additional π-stacking distance is observed, indicating the insertion of two PIPER ligands into the tail-to-tail arrangement of G-quadruplex dimer. DEER technology can reveal a new binding mode of G-quadruplex binder PIPER forming a sandwich complex with the G-quadruplex dimer.
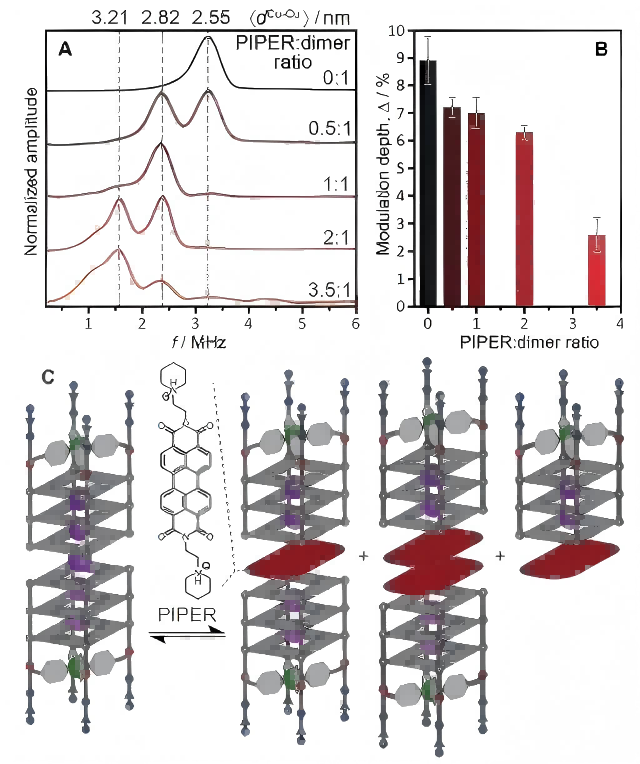
Figure 3 (A) DEER dipolar spectra (geff = 2.061) of [Cu2+@A4]2 dimers with different ratios of PIPER; (B) DEER modulation depths of [Cu2+@A4]2 dimers with different ratios of PIPER; (C) Equilibrium between [Cu2+@A4]2 dimers and PIPER@[Cu2+@A4]2, 2PIPER@[Cu2+@A4]2, PIPER@[Cu2+@A4]. (Angew. Chem. Int. Ed. 2021, 60, 4939–4947)
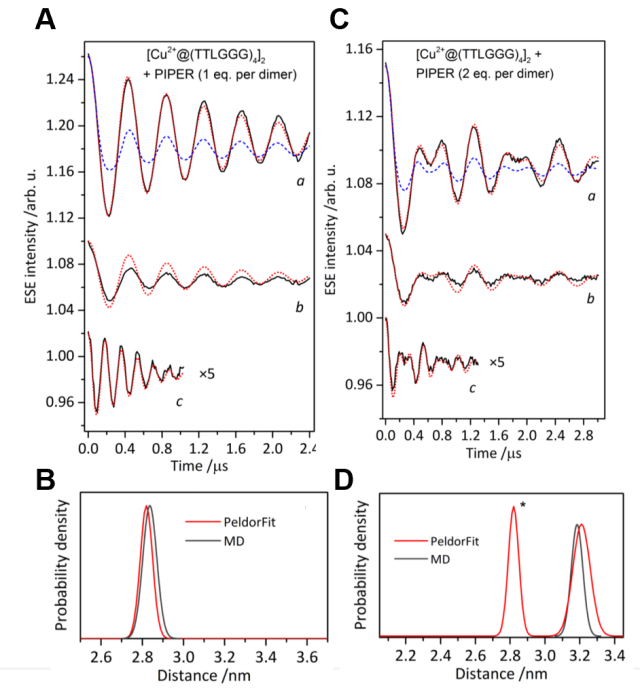
Figure 4 (A) DEER time-domain spectra of PIPER@[Cu2+@A4]2; (B) Distance distribution of PIPER@[Cu2+@A4]2 obtained by PeldorFit (red line) and MD simulation (gray line); (C) DEER time-domain spectra of 2PIPER@[Cu2+@A4]2; (D) Distance distribution of 2PIPER@[Cu2+@A4]2 obtained by PeldorFit (red line) and MD simulation (gray line). (Angew. Chem. Int. Ed. 2021, 60, 4939–4947)
CIQTEK Pulsed Electron Paramagnetic Resonance (EPR) Spectrometer
The EPR100 Pulsed Electron Paramagnetic Resonance (EPR) spectrometer from CIQTEK is capable of electron-electron double resonance (DEER) techniques, which can be used to study the structure, function, physiological processes, and mechanisms of protein molecules that play a critical role in various diseases, including complex membrane proteins, DNA, RNA, and nucleoprotein complexes.

CIQTEK X-band Pulsed Electron Paramagnetic Resonance (EPR) Spectrometer EPR100
Experimental results processed using DeerAnalysis
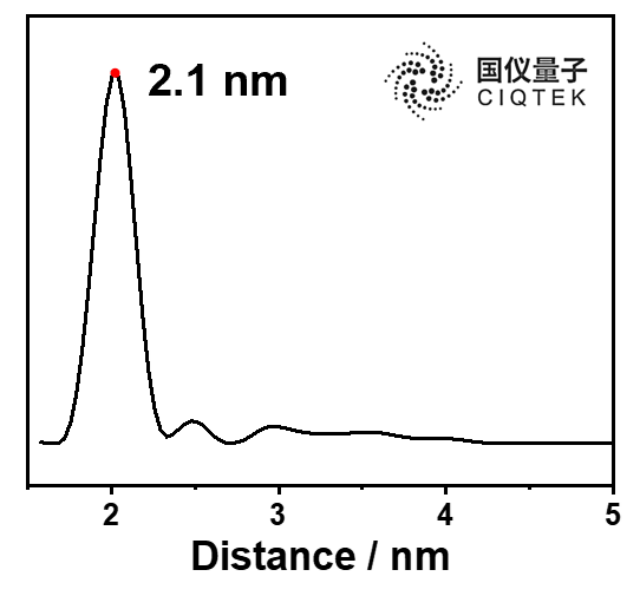
The DEER experimental results obtained using CIQTEK’s EPR100
Overall, the application of electron-electron double resonance (DEER) techniques in conjunction with site-directed spin labeling (SDSL) and the use of CIQTEK’s EPR100 spectrometer provide valuable insights into the structure and binding modes of G-quadruplexes, as well as the interaction between G-quadruplex binders and dimers. These findings contribute to the understanding of cancer biology and the development of potential anticancer drugs.